VITIS: Vol. 63, Art. 6, 8 pp. (2024) | DOI: 10.5073/vitis.2024.63.06. | Kaur et al.
Exogenous application of double-stranded RNA to reduce grapevine Pinot gris virus titre in in vitro grown Vitis vinifera
 | (c) The author(s) 2024 This is an Open Access article distributed under the terms of the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/deed.en). |
Submitted/accepted for publication: March, 3, 2024./June, 17, 2024. |
Table S1: Contents of initiation media to prepare grapevine tissue culture plantlets.
Ingredient | Amount |
Murashige and Skoog (MS) Salts | 2.2 g/L |
Indole-3-butyric acid (IBA) | 0.3 mg/L |
saccharose | 25 g/L |
agar | 7 g/L |
Table S2: The GenBank accession numbers for amino acid (aa) sequences of the RNA-dependent RNA polymerase (RdRp), Movement Protein (MP) and Coat Protein (CP) for six exemplar species in the Trichovirus genus including Grapevine Pinot virus, Apple chlorotic leaf spot virus, Apricot pseudo-chlorotic leaf spot virus, Cherry mottle leaf virus, Peach mosaic virus and Grapevine berry inner necrosis virus.
Trichovirus species | RNA-dependent RNA polymerase (RdRp) GenBank accession number | Movement Protein (MP) GenBank accession number | Coat Protein (CP) GenBank accession number |
Apple chlorotic leaf spot virus | NP_040551.1 | NP_040552.1 | NP_040553.1 |
Apricot pseudo-chlorotic leaf spot virus | YP_224130.1 | YP_224131.1 | YP_224132.1 |
Cherry mottle leaf virus | NP_062428.1 | NP_062429.1 | NP_062430.1 |
Peach mosaic virus | YP_002308565.1 | YP_002308566.1 | YP_002308567.1 |
Grapevine berry inner necrosis virus | YP_004293216.1 | YP_004293217.1 | YP_004293218.1 |
Grapevine Pinot gris virus | YP_004732978.2 | YP_004732979.2 | YP_004732980.2 |
Table S3: The GenBank accession numbers for amino acid (aa) sequences of the RNA-dependent RNA polymerase (RdRp), Movement Protein (MP) and Coat Protein (CP) for 200 grapevine Pinot gris virus (GPGV) genome sequences and their respective country of origin.
Isolate | RdRp | MP | CP | Isolate | RdRp | MP | CP | Isolate | RdRp | MP | CP |
ERR922628-GPGV | DAC84876 | DAC84877 | DAC84878 | Beb3-GPGV2 | QFG15452 | QFG15453 | QFG15454 | CK1 | WCZ55034 | WCZ55035 | WCZ55036 |
ERR922630-GPGV1 | DAC84879 | DAC84880 | DAC84881 | Beb8-GPGV | QFG15455 | QFG15456 | QFG15457 | 2.1 | WCZ55037 | WCZ55038 | WCZ55039 |
ERR922630-GPGV2 | DAC84882 | DAC84883 | DAC84884 | Ma17-10-14-GPGV | QFG15458 | QFG15459 | QFG15460 | 2.12 | WCZ55040 | WCZ55041 | WCZ55042 |
ERR922630-GPGV3 | DAC84885 | DAC84886 | DAC84887 | Ma17-10-18-GPGV1 | QFG15461 | QFG15462 | QFG15463 | 2.17 | WCZ55043 | WCZ55044 | WCZ55045 |
ERR922631-GPGV | DAC84888 | DAC84889 | DAC84890 | Ma17-10-18-GPGV2 | QFG15464 | QFG15465 | QFG15466 | 5.5 | WCZ55046 | WCZ55047 | WCZ55048 |
ERR922632-GPGV1 | DAC84891 | DAC84892 | DAC84893 | Ma17-12-7-GPGV | QFG15467 | QFG15468 | QFG15469 | 5.6 | WCZ55049 | WCZ55050 | WCZ55051 |
ERR922632-GPGV2 | DAC84894 | DAC84895 | DAC84896 | Ma17-2-27-GPGV | QFG15470 | QFG15471 | QFG15472 | 5.13 | WCZ55052 | WCZ55053 | WCZ55054 |
ERR922633-GPGV1 | DAC84897 | DAC84898 | DAC84899 | Ma17-3-24-GPGV | QFG15473 | QFG15474 | QFG15475 | 5.14 | WCZ55055 | WCZ55056 | WCZ55057 |
ERR922633-GPGV2 | DAC84900 | DAC84901 | DAC84902 | Ma17-3-26-GPGV1 | QFG15476 | QFG15477 | QFG15478 | 5.17 | WCZ55058 | WCZ55059 | WCZ55060 |
ERR923264-GPGV2 | DAC84903 | DAC84904 | DAC84905 | Ma17-3-26-GPGV2 | QFG15479 | QFG15480 | QFG15481 | 5.21 | WCZ55061 | WCZ55062 | WCZ55063 |
ERR923264-GPGV1 | DAC84906 | DAC84907 | DAC84908 | Ma17-3-27-GPGV1 | QFG15482 | QFG15483 | QFG15484 | 5.22 | WCZ55064 | WCZ55065 | WCZ55066 |
ERR926756-GPGV | DAC84909 | DAC84910 | DAC84911 | Ma17-3-27-GPGV2 | QFG15485 | QFG15486 | QFG15487 | 5.24 | WCZ55067 | WCZ55068 | WCZ55069 |
SRR1631863-67-68-GPGV | DAC84912 | DAC84913 | DAC84914 | Ma17-3-35-GPGV1 | QFG15488 | QFG15489 | QFG15490 | 8.6 | WCZ55070 | WCZ55071 | WCZ55072 |
SRR1658425-26-27-GPGV1 | DAC84915 | DAC84916 | DAC84917 | Ma17-5-36-GPGV1 | QFG15491 | QFG15492 | QFG15493 | 8.7 | WCZ55073 | WCZ55074 | WCZ55075 |
SRR1658425-26-27-GPGV2 | DAC84918 | DAC84919 | DAC84920 | Ma17-5-36-GPGV2 | QFG15494 | QFG15495 | QFG15496 | 8.28 | WCZ55076 | WCZ55077 | WCZ55078 |
SRR2120794-GPGV | DAC84921 | DAC84922 | DAC84923 | Ma17-A2-37-GPGV | QFG15497 | QFG15498 | QFG15499 | 8.29 | WCZ55079 | WCZ55080 | WCZ55081 |
SRR2845691-GPGV | DAC84924 | DAC84925 | DAC84926 | Ma17-A4-24-GPGV1 | QFG15500 | QFG15501 | QFG15502 | 8.33 | WCZ55082 | WCZ55083 | WCZ55084 |
SRR3167555-GPGV | DAC84927 | DAC84928 | DAC84929 | Ma17-A4-24-GPGV2 | QFG15503 | QFG15504 | QFG15505 | 8.38 | WCZ55085 | WCZ55086 | WCZ55087 |
SRR3190105-GPGV | DAC84930 | DAC84931 | DAC84932 | Ma17-A4-25-GPGV | QFG15506 | QFG15507 | QFG15508 | 8.47 | WCZ55088 | WCZ55089 | WCZ55090 |
SRR3288835-GPGV | DAC84933 | DAC84934 | DAC84935 | Ma17-A4-25-GPGV | QFG15509 | QFG15510 | QFG15511 | LT6 | WCZ55091 | WCZ55092 | WCZ55093 |
SRR3288835-GPGV | DAC84936 | DAC84937 | DAC84938 | Ma17-A8-12-GPGV | QFG15512 | QFG15513 | QFG15514 | LT7 | WCZ55094 | WCZ55095 | WCZ55096 |
SRR5227657-GPGV | DAC84939 | DAC84940 | DAC84941 | Ma17-A8-20-GPGV | QFG15515 | QFG15516 | QFG15517 | 9.1 | WCZ55097 | WCZ55098 | WCZ55099 |
SRR5332103-GPGV1 | DAC84942 | DAC84943 | DAC84944 | Ma17-A9-20-GPGV1 | QFG15518 | QFG15519 | QFG15520 | 9.2 | WCZ55100 | WCZ55101 | WCZ55102 |
SRR5332103-GPGV2 | DAC84945 | DAC84946 | DAC84947 | Ma17-A9-20-GPGV2 | QFG15521 | QFG15522 | QFG15523 | 9.3 | WCZ55103 | WCZ55104 | WCZ55105 |
SRR5332104-GPGV1 | DAC84948 | DAC84949 | DAC84950 | Ma17-A9-20-GPGV2 | QFG15524 | QFG15525 | QFG15526 | 9.4 | WCZ55106 | WCZ55107 | WCZ55108 |
SRR5332104-GPGV2 | DAC84951 | DAC84952 | DAC84953 | Ma17-A9-21-GPGV2 | QFG15527 | QFG15528 | QFG15529 | 9.5 | WCZ55109 | WCZ55110 | WCZ55111 |
SRR5332104-GPGV3 | DAC84954 | DAC84955 | DAC84956 | Ma17-A9-22-GPGV | QFG15530 | QFG15531 | QFG15532 | 9.6 | WCZ55112 | WCZ55113 | WCZ55114 |
SRR5332107-GPGV | DAC84957 | DAC84958 | DAC84959 | GPgV_MID38_ | QFG15533 | QFG15534 | QFG15535 | 9.9 | WCZ55115 | WCZ55116 | WCZ55117 |
SRR5332108-GPGV | DAC84960 | DAC84961 | DAC84962 | Pa10-GPGV | QFG15536 | QFG15537 | QFG15538 | 9.1 | WCZ55118 | WCZ55119 | WCZ55120 |
SRR5457616-GPGV | DAC84963 | DAC84964 | DAC84965 | IFV72_Vau_A224 | QFG15539 | QFG15540 | QFG15541 | 9.11 | WCZ55121 | WCZ55122 | WCZ55123 |
SRR5457630-GPGV | DAC84966 | DAC84967 | DAC84968 | IP-C522-GPGV1 | QFG15542 | QFG15543 | QFG15544 | 9.13 | WCZ55124 | WCZ55125 | WCZ55126 |
SRR5457635-GPGV | DAC84969 | DAC84970 | DAC84971 | IP-C522-GPGV2 | QFG15545 | QFG15546 | QFG15547 | 9.14 | WCZ55127 | WCZ55128 | WCZ55129 |
SRR5457636-GPGV | DAC84972 | DAC84973 | DAC84974 | IV2_I19-12 | QFG15548 | QFG15549 | QFG15550 | Riesling 25-3 | AOG16120 | AOG16121 | AOG16122 |
SRR5457637-GPGV | DAC84975 | DAC84976 | DAC84977 | IV4_I68-2-11 | QFG15551 | QFG15552 | QFG15553 | PN | ASA45580 | ASA45581 | ASA45582 |
SRR5457659-GPGV | DAC84978 | DAC84979 | DAC84980 | IV5_I76-1 | QFG15554 | QFG15555 | QFG15556 | SL13 | AVD53915 | AVD53916 | AVD53917 |
SRR5457660-GPGV | DAC84981 | DAC84982 | DAC84983 | IV6_I69-4-2 | QFG15557 | QFG15558 | QFG15559 | SL53 | AVD53918 | AVD53919 | AVD53920 |
SRR5457661-GPGV | DAC84984 | DAC84985 | DAC84986 | IV6_I70-5-3 | QFG15560 | QFG15561 | QFG15562 | H-JP1 | BCR39156 | BCR39157 | BCR39158 |
SRR5457662-GPGV-1 | DAC84987 | DAC84988 | DAC84989 | IV6_I70-5-3 | QFG15563 | QFG15564 | QFG15565 | H-JP2 | BCR39159 | BCR39160 | BCR39161 |
SRR5457662-GPGV-2 | DAC84990 | DAC84991 | DAC84992 | IV7_F81 | QFG15566 | QFG15567 | QFG15568 | fvg-Is1 | AYN70038 | AYN70039 | AYN70040 |
SRR5457663-GPGV-1 | DAC84993 | DAC84994 | DAC84995 | IV7_I78-2 | QFG15569 | QFG15570 | QFG15571 | fvg-ls6 | AYN70041 |
|
|
SRR5457663-GPGV-2 | DAC84996 | DAC84997 | DAC84998 | IV8_F82 | QFG15572 | QFG15573 | QFG15574 | fvg-ls7 | AYN70042 |
|
|
SRR5457664-GPGV | DAC84999 | DAC85000 | DAC85001 | IV8_F85 | QFG15575 | QFG15576 | QFG15577 | Fvg-ls8 | AYN70043 |
|
|
SRR7275242-GPGV | DAC85002 | DAC85003 | DAC85004 | IV9_F83-2 | QFG15578 | QFG15579 | QFG15580 | Fvg-ls12 | AYN70044 | AYN70045 |
|
SRR7275243-GPGV | DAC85005 | DAC85006 | DAC85007 | W22-GPGV1 | QFG15581 | QFG15582 | QFG15583 | Fvg-ls13 | AYN70046 |
|
|
SRR8260939-GPGV | DAC85008 | DAC85009 | DAC85010 | W22-GPGV2 | QFG15584 | QFG15585 | QFG15586 | Fvg-ls14 | AYN70047 |
|
|
SRR8260948-GPGV-1 | DAC85011 | DAC85012 | DAC85013 | W25-GPGV | QFG15587 | QFG15588 | QFG15589 | Fvg-ls15 | AYN70048 | AYN70049 |
|
SRR8260948-GPGV-2 | DAC85014 | DAC85015 | DAC85016 | GPGV-Gr- | QUS52473 | QUS52474 | QUS52475 | Fvg-ls17 | AYN70050 | AYN70051 |
|
SRR8260950-GPGV | DAC85017 | DAC85018 | DAC85019 | GPGV-Gr- | QUS52490 | QUS52491 | QUS52492 | 12G1110 | QBZ78363 | QBZ78364 | QBZ78365 |
SRR866574-GPGV | DAC85020 | DAC85021 | DAC85022 | 12G4110 | UCJ00541 | UCJ00542 | UCJ00543 | S103 | QEQ49999 | QEQ50000 | QEQ50001 |
SRR866575-GPGV | DAC85023 | DAC85024 | DAC85025 | 12G4111 | UCJ00544 |
|
| S104 | QEQ50002 | QEQ50003 | QEQ50004 |
SRR866575-GPGV | CCC20963 | CCC20964 | CCC20965 | 13C233 | UCJ00556 |
|
| S105 | QEQ50005 | QEQ50006 | QEQ50007 |
SK30 | AGV76021 | AGV76022 | AGV76023 | Rk3 | UVC46710 | UVC46711 | UVC46712 | S106 | QEQ50008 | QEQ50009 | QEQ50010 |
SK01 | AGV76024 | AGV76025 | AGV76026 | GPGV_CH_M | UTM04226 | UTM04227 | UTM04228 | S123 | QEQ50011 | QEQ50012 | QEQ50013 |
SK13 | AGV76027 | AGV76028 | AGV76029 | A1575h | UTH78988 | UTH78989 | UTH78990 | S107 | QEQ50014 | QEQ50015 | QEQ50016 |
SK30-1 | AID59467 | AID59468 | AID59469 | A1868h | UTH78991 | UTH78992 | UTH78993 | S111 | QEQ50017 | QEQ50018 | QEQ50019 |
Mer | AIY29054 | AIY29055 | AIY29056 | A1873h | UTH78994 | UTH78995 | UTH78996 | S115 | QEQ50020 | QEQ50021 | QEQ50022 |
TI_21 | AKP16327 |
|
| A1542h | UTH78997 | UTH78998 | UTH78999 | S142 | QEQ50023 | QEQ50024 | QEQ50025 |
TI_25 | AKP16328 |
|
| A1872h | UTH79000 | UTH79001 | UTH79002 | S145 | QEQ50026 | QEQ50027 | QEQ50028 |
7_2_5 | AKP16329 |
|
| A1581h | UTH79003 | UTH79004 | UTH79005 | S147 | QEQ50029 | QEQ50030 | QEQ50031 |
20_5_3 | AKP16330 |
|
| A1584h | UTH79006 | UTH79007 | UTH79008 | S148 | QEQ50032 | QEQ50033 | QEQ50034 |
22_2_3 | AKP16331 |
|
| A1892h | UTH79011 |
|
| GPGV-136-NS-1 | QKV50564 | QKV50565 | QKV50566 |
GPGV-TN | AMQ49210 | AMQ49211 | AMQ49212 | A1881h | UVC46658 | UVC46659 | UVC46660 | Regent-BE | QIE48965 | QIE48966 | QIE48967 |
BC-1 | AML83897 | AML83898 | AML83899 | A1574h | UVC46661 | UVC46662 | UVC46663 | Beb13-GPGV1 | QFG15440 | QFG15441 | QFG15442 |
GPGV_FEM01 | ANQ87152 | ANQ87153 | ANQ87154 | A1588h | UVC46664 | UVC46665 |
| Beb13-GPGV2 | QFG15443 | QFG15444 | QFG15445 |
Goldfinger | APT42884 | APT42885 | APT42886 | A1889h | UVC46666 | UVC46667 | UVC46668 | Beb16-GPGV | QFG15446 | QFG15447 | QFG15448 |
SK704 | ANA05793 | ANA05792 | ANA05791 | A1890h | UVC46669 | UVC46670 | UVC46671 | Beb3-GPGV1 | QFG15449 | QFG15450 | QFG15451 |
Table S4: The sample identity, cultivar, RT-qPCR cycle threshold value (Ct), number of raw reads generated from metagenomic high-throughput sequencing (HTS), number of reads after quality trimming, number of contigs generated by SPAdes, number of viral contigs, viral contig size, its percentage nucleotide identity to the closest matching isolate, average mapped read depth, and number of mapped reads calculated using Geneious in the grapevine sample.
Sample Identity | Cultivar | Ct value | Total number of raw reads | Reads after trimming | No of viral contigs | Assembled GPGV contig size (nt) | Percentage | Average mapped read depth | Number of mapped reads |
Grafted grapevine | Sauvignon blanc | 18.6 | 2215609 | 2209873 | 10 (GPGV) | 7275 | 99.4 (to NCBI | 22 | 785 |
2 (GRSPaV*) | 8681 | 99.7 (to NCBI GenBank isolate NC_001948.1) | 54 | 2647 |
*GPGV: Grapevine Pinot gris virus
*GRSPaV: Grapevine rupestris stem pitting associated virus
Table S5: Conserved amino acid sequence motifs occurring in order as they are encoded along the grapevine Pinot gris virus (GPGV) genome, 5’ to 3’, in the RNA-dependent RNA polymerase (RdRp), Movement Protein (MP) and Coat Protein (CP) of the six exemplar species of the genus Trichovirus including Grapevine Pinot virus, Apple chlorotic leaf spot virus, Apricot pseudo-chlorotic leaf spot virus, Cherry mottle leaf virus, Peach mosaic virus and Grapevine berry inner necrosis virus.
ORF region Conserved Trichovirus motifs (function) | Conserved amino acid motifs in the exemplar Trichovirus species | ||
RNA-dependent RNA polymerase (RdRp) | Movement Protein (MP) | Coat Protein (CP) | |
1 | YRTP | PI | NIAV* |
2 | EEL | SS | GTS* |
3 | LSP | FR | FDFN |
4 | HSHP | NY | IF* |
5 | CKTLEN | GALSISID | RQ* |
6 | SIK | LF | FA* |
7 | NRLV | GV | GL |
8 | RYG* | RW | PE* |
9 | HDE | LQ |
|
10 | VFP* | SPN |
|
11 | EIL | DP |
|
12 | PDG* | LNF |
|
13 | FGPYD* | SVR |
|
14 | DVG | FG |
|
15 | IYVRVPI (GKVRVPI) |
|
|
16# | FKKI* |
|
|
17# | SLKKPD* |
|
|
18# | AKLRQL* |
|
|
19# | DDF* |
|
|
20# | TLRV* |
|
|
21 | KLLK |
|
|
22 | TGI |
|
|
23# | FGFAGSGKSH* |
|
|
24# | TFESALK* |
|
|
25# | GDPLQA* |
|
|
26 | LVAS |
|
|
27 | GESQGLTF |
|
|
28 | LSEE* |
|
|
29 | LCSD* |
|
|
30 | TRF |
|
|
31 | WLK* |
|
|
32 | REFK |
|
|
33 | KKRLRF |
|
|
34 | RSD* |
|
|
35 | DW* |
|
|
36 | KSQLCTKFEKRF |
|
|
37 | AKAGQTLACF |
|
|
38 | FSPWCRY |
|
|
39# | HQRK* |
|
|
40# | ESDY* |
|
|
41# | AFDVSQDH* |
|
|
42# | FEV* |
|
|
43# | LGC* |
|
|
44# | FAIMRFTGEF* |
|
|
45# | TFL* |
|
|
46# | GDDMCAL* |
|
|
47# | LKAKV* |
|
|
48 | FCGWRL |
|
|
49 | KEP |
|
|
50 | ERL |
|
|
51 | LEF |
|
|
52 | AYKLG |
|
|
53 | RFF |
|
|
54 | HLL |
|
|
55 | RGYSN |
|
|
* Conserved motifs in RdRp and CP of the GPGV genome in both aa sequence and position have been marked in bold.
# The three RdRp regions with five (FKKI, SLKKPD, AKLRQL, DDF and TLRV); three (FGFAGSGKS, TFESA, GDPLQA) and nine (HQRK, ESDY, AFDVSQDH, FEV, LGC, FAIMRFTGEF, TFL, GDDMCAL and LKAKV) conserved amino acid motifs, respectively, have been highlighted
Table S6: The cycle threshold (Ct) values with standard deviation before and after seven days post application of four treatments (nine replicates and three technical replicates for each) including a short synthetic grapevine Pinot gris virus (GPGV) targeted dsRNA, second PCR product, RNase-free water and no application to tissue culture (TC) grapevine shoot tips for 2 or 24 hours prior to reintroduction into TC or directly applying treatments to the leaves of plantlets in TC.
Application 1- Dipping excised shoot tips for 2 hrs and then introduce in TC plant culture vessels | |||||||||
| Before application | dip in dsRNA for 2 hrs | Difference in Ct | ||||||
| Technical replicate 1 | Technical replicate 2 | Technical replicate 3 | Technical replicate 1 | Technical replicate 2 | Technical replicate 3 | Technical replicate 1 | Technical replicate 2 | Technical replicate 3 |
replicate 1 | 20.9 | 21.2 | 21.2 | 23.5 | 23.5 | 23.7 | 2.6 | 2.3 | 2.5 |
replicate 2 | 20.9 | 21.2 | 21.2 | 23.8 | 24.3 | 24.2 | 2.9 | 3.1 | 2.9 |
replicate 3 | 20.9 | 21.2 | 21.2 | 23.3 | 23.5 | 23.8 | 2.4 | 2.3 | 2.6 |
replicate 4 | 20.9 | 21.2 | 21.2 | 24.5 | 24.8 | 25.0 | 3.5 | 3.6 | 3.8 |
replicate 5 | 20.9 | 21.2 | 21.2 | 24.0 | 23.6 | 23.9 | 3.1 | 2.4 | 2.7 |
replicate 6 | 20.9 | 21.2 | 21.2 | 23.9 | 23.3 | 23.1 | 3.0 | 2.1 | 1.8 |
replicate 7 | 20.9 | 21.2 | 21.2 | 22.7 | 22.8 | 22.3 | 1.8 | 1.6 | 1.0 |
replicate 8 | 20.9 | 21.2 | 21.2 | 23.6 | 23.6 | 24.0 | 2.6 | 2.4 | 2.8 |
replicate 9 | 20.9 | 21.2 | 21.2 | 25.5 | 26.1 | 25.7 | 4.6 | 4.9 | 4.4 |
| Average = 21.1 | Standard deviation = 0.1 | Average = 23.9 | Standard deviation = 0.9 | Average = 2.8 | Standard deviation = 0.9 | Significance level (P) < 0.0001 | ||
replicate 1 | 19.6 | 19.8 | 19.7 | 19.8 | 20.2 | 20.2 | 19.9 | 20.2 | 20.2 |
replicate 2 | 19.6 | 19.8 | 19.7 | 20.8 | 20.9 | 20.2 | 20.5 | 20.3 | 20.4 |
replicate 3 | 19.6 | 19.8 | 19.7 | 20.5 | 21.0 | 20.2 | 20.2 | 21.2 | 20.2 |
replicate 4 | 19.6 | 19.8 | 19.7 | 20.9 | 21.3 | 21.5 | 20.9 | 21.3 | 20.8 |
replicate 5 | 19.6 | 19.8 | 19.7 | 19.5 | 19.2 | 19.1 | 19.5 | 19.2 | 19.8 |
replicate 6 | 19.6 | 19.8 | 19.7 | 19.0 | 19.3 | 19.2 | 19.5 | 19.7 | 19.7 |
replicate 7 | 19.6 | 19.8 | 19.7 | 21.0 | 21.1 | 21.1 | 20.4 | 20.3 | 20.6 |
replicate 8 | 19.6 | 19.8 | 19.7 | 20.5 | 20.6 | 20.7 | 20.5 | 20.6 | 20.3 |
replicate 9 | 19.6 | 19.8 | 19.7 | 20.1 | 20.0 | 20.6 | 20.1 | 20.2 | 19.8 |
| Average = 19.7 | Standard deviation = 0.1 | Average = 20.2 | Standard deviation = 0.5 | Average = 0.5 | Standard deviation = 0.5 | Significance level (P) 0.10 > P > 0.05 | ||
| Before application | dip in buffer control (water) for 2 hrs | Difference in Ct | ||||||
| Technical replicate 1 | Technical replicate 2 | Technical replicate 3 | Technical replicate 1 | Technical replicate 2 | Technical replicate 3 | Technical replicate 1 | Technical replicate 2 | Technical |
replicate 1 | 20.9 | 20.7 | 20.8 | 21.5 | 21.7 | 22 | 0.6 | 1.0 | 1.2 |
replicate 2 | 20.9 | 20.7 | 20.8 | 21.2 | 21.6 | 21.4 | 0.3 | 0.9 | 0.6 |
replicate 3 | 20.9 | 20.7 | 20.8 | 21.1 | 21.4 | 21.7 | 0.2 | 0.7 | 0.9 |
replicate 4 | 20.9 | 20.7 | 20.8 | 21.0 | 21.3 | 21.9 | 0.1 | 0.6 | 1.1 |
replicate 5 | 20.9 | 20.7 | 20.8 | 21.6 | 21.5 | 22.0 | 0.7 | 0.8 | 1.2 |
replicate 6 | 20.9 | 20.7 | 20.8 | 21.0 | 21.6 | 20.6 | 0.1 | 0.9 | -0.2 |
replicate 7 | 20.9 | 20.7 | 20.8 | 20.8 | 20.9 | 21.0 | -0.1 | 0.2 | 0.2 |
replicate 8 | 20.9 | 20.7 | 20.8 | 21.1 | 20.8 | 20.7 | 0.2 | 0.1 | -0.1 |
replicate 9 | 20.9 | 20.7 | 20.8 | 20.7 | 20.5 | 20.9 | -0.2 | -0.2 | 0.1 |
| Average = 20.8 | Standard deviation = 0.1 | Average = 21.2 | Standard deviation = 0.4 | Average = 0.4 | Standard deviation = 0.5 | Significance level (P) 0.10 > P > 0.05 | ||
| Before application | no treatment | Difference in Ct | ||||||
| Technical replicate 1 | Technical replicate 2 | Technical replicate 3 | Technical replicate 1 | Technical replicate 2 | Technical replicate 3 | Technical replicate 1 | Technical replicate 2 | Technical |
replicate 1 | 21.7 | 21.8 | 21.9 | 22.0 | 22.6 | 22.2 | 0.4 | 0.8 | 0.4 |
replicate 2 | 21.7 | 21.8 | 21.9 | 22.5 | 22.5 | 22.8 | 0.9 | 0.6 | 1.0 |
replicate 3 | 21.7 | 21.8 | 21.9 | 22.3 | 22.2 | 22.6 | 0.7 | 0.4 | 0.7 |
replicate 4 | 21.7 | 21.8 | 21.9 | 21.5 | 21.3 | 21.6 | -0.2 | -0.5 | -0.3 |
replicate 5 | 21.7 | 21.8 | 21.9 | 20.6 | 20.6 | 20.8 | -1.0 | -1.2 | -1.1 |
replicate 6 | 21.7 | 21.8 | 21.9 | 21.2 | 21.2 | 21.4 | -0.5 | -0.7 | -0.4 |
replicate 7 | 21.7 | 21.8 | 21.9 | 20.6 | 20.3 | 20.7 | -1.1 | -1.5 | -1.2 |
replicate 8 | 21.7 | 21.8 | 21.9 | 22.7 | 22.7 | 23.1 | 1.0 | 0.9 | 1.2 |
replicate 9 | 21.7 | 21.8 | 21.9 | 22.2 | 21.9 | 22.5 | 0.6 | 0.1 | 0.6 |
| Average = 21.8 | Standard deviation = 0.1 | Average = 21.8 | Standard deviation = 0.8 | Average = -0.0 | Standard deviation = 0.8 | Significance level (P) > 0.10 (P= 1.0000) | ||
replicate 1 | 19.6 | 19.8 | 20.0 | 24.1 | 23 | 23.4 | 4.5 | 3.2 | 3.4 |
replicate 2 | 19.6 | 19.8 | 20.0 | 25.5 | 25.7 | 25.8 | 6.0 | 5.9 | 5.8 |
replicate 3 | 19.6 | 19.8 | 20.0 | 26.2 | 26.8 | 27.1 | 6.7 | 7.0 | 7.1 |
replicate 4 | 19.6 | 19.8 | 20.0 | 24.1 | 23.7 | 25.0 | 4.5 | 4.0 | 5.0 |
replicate 5 | 19.6 | 19.8 | 20.0 | 24.1 | 24.7 | 25.0 | 4.5 | 5.0 | 5.0 |
replicate 6 | 19.6 | 19.8 | 20.0 | 23.3 | 23.7 | 23.9 | 3.7 | 3.9 | 3.9 |
replicate 7 | 19.6 | 19.8 | 20.0 | 24.9 | 24.7 | 24.9 | 5.3 | 5.0 | 5.0 |
replicate 8 | 19.6 | 19.8 | 20.0 | 26.1 | 26.4 | 26.8 | 6.5 | 6.7 | 6.9 |
replicate 9 | 19.6 | 19.8 | 20.0 | 24.2 | 24 | 24.4 | 4.6 | 4.2 | 4.4 |
| Average = 19.8 | Standard deviation = 0.1 | Average = 24.9 | Standard deviation = 1.2 | Average = 5.1 | Standard deviation = 1.2 | Significance level (P) < 0.0001 | ||
| Before application | dip in the second PCR product for 24 hrs | Difference in Ct | ||||||
| Technical replicate 1 | Technical replicate 2 | Technical replicate 3 | Technical replicate 1 | Technical replicate 2 | Technical replicate 3 | Technical replicate 1 | Technical replicate 2 | Technical |
replicate 1 | 21.2 | 21.6 | 21.6 | 21.7 | 21.9 | 22.1 | 0.5 | 0.4 | 0.5 |
replicate 2 | 21.2 | 21.6 | 21.6 | 21.8 | 21.1 | 22.2 | 0.6 | -0.5 | 0.6 |
replicate 3 | 21.2 | 21.6 | 21.6 | 21.6 | 21.2 | 22.4 | 0.4 | -0.4 | 0.8 |
replicate 4 | 21.2 | 21.6 | 21.6 | 21.5 | 22.2 | 22.4 | 0.3 | 0.6 | 0.7 |
replicate 5 | 21.2 | 21.6 | 21.6 | 22.0 | 21.8 | 21.9 | 0.8 | 0.3 | 0.2 |
replicate 6 | 21.2 | 21.6 | 21.6 | 20.8 | 20.3 | 21.7 | -0.3 | -1.3 | 0.1 |
replicate 7 | 21.2 | 21.6 | 21.6 | 21.5 | 21.4 | 22.6 | 0.3 | -0.2 | 1.0 |
replicate 8 | 21.2 | 21.6 | 21.6 | 22.2 | 21.2 | 22.4 | 1.0 | -0.3 | 0.8 |
replicate 9 | 21.2 | 21.6 | 21.6 | 20.7 | 20.9 | 21.0 | -0.4 | -0.7 | -0.7 |
| Average = 21.5 | Standard deviation = 0.2 | Average = 21.7 | Standard deviation = 0.6 | Average = 0.2 | Standard deviation = 0.6 | Significance level (P) > 0.10 | ||
| Before application | dip in buffer control (water) for 24 hrs | Difference in Ct | ||||||
| Technical replicate 1 | Technical replicate 2 | Technical replicate 3 | Technical replicate 1 | Technical replicate 2 | Technical replicate 3 | Technical replicate 1 | Technical replicate 2 | Technical |
replicate 1 | 20.0 | 19.1 | 19.2 | 20.3 | 19.6 | 19.4 | 0.3 | 0.5 | 0.2 |
replicate 2 | 20.0 | 19.1 | 19.2 | 20.0 | 19.4 | 20.2 | 0.0 | 0.2 | 1.1 |
replicate 3 | 20.0 | 19.1 | 19.2 | 20.5 | 19.7 | 20.8 | 0.5 | 0.6 | 1.6 |
replicate 4 | 20.0 | 19.1 | 19.2 | 19.6 | 19.9 | 18.9 | -0.4 | 0.8 | -0.3 |
replicate 5 | 20.0 | 19.1 | 19.2 | 20.1 | 19.4 | 20.2 | 0.1 | 0.3 | 1.0 |
replicate 6 | 20.0 | 19.1 | 19.2 | 20.7 | 19.8 | 19.6 | 0.7 | 0.6 | 0.4 |
replicate 7 | 20.0 | 19.1 | 19.2 | 19.7 | 18.7 | 19.6 | -0.2 | -0.4 | 0.5 |
replicate 8 | 20.0 | 19.1 | 19.2 | 20.0 | 19.0 | 18.9 | 0.0 | -0.1 | -0.3 |
replicate 9 | 20.0 | 19.1 | 19.2 | 20.1 | 19.3 | 20.0 | 0.2 | 0.2 | 0.8 |
| Average = 19.4 | Standard deviation = 0.4 | Average = 19.8 | Standard deviation = 0.5 | Average = 0.4 | Standard deviation = 0.5 | Significance level (P) 0.10 > P > 0.05 | ||
replicate 1 | 20.6 | 20.8 | 21.0 | 21.1 | 21.5 | 22 | 0.5 | 0.6 | 1.0 |
replicate 2 | 20.6 | 20.8 | 21.0 | 20.7 | 21.2 | 21.6 | 0.1 | 0.4 | 0.6 |
replicate 3 | 20.6 | 20.8 | 21.0 | 21.1 | 20.5 | 21.6 | 0.5 | -0.3 | 0.6 |
replicate 4 | 20.6 | 20.8 | 21.0 | 20.9 | 20.5 | 21.1 | 0.3 | -0.3 | 0.2 |
replicate 5 | 20.6 | 20.8 | 21.0 | 20.6 | 21.0 | 21.9 | 0.0 | 0.2 | 0.9 |
replicate 6 | 20.6 | 20.8 | 21.0 | 20.4 | 20.7 | 20.4 | -0.2 | -0.2 | -0.5 |
replicate 7 | 20.6 | 20.8 | 21.0 | 20.7 | 21.0 | 20.7 | 0.1 | 0.1 | -0.2 |
replicate 8 | 20.6 | 20.8 | 21.0 | 21.5 | 20.5 | 21.2 | 0.9 | -0.4 | 0.2 |
replicate 9 | 20.6 | 20.8 | 21.0 | 20.6 | 19.6 | 20.6 | 0.0 | -1.3 | -0.4 |
| Average = 20.8 | Standard deviation = 0.2 | Average = 20.9 | Standard deviation = 0.5 | Average = 0.1 | Standard deviation = 0.5 | Significance level (P) > 0.10 | ||
Application 3- Direct application of dsRNA in TC plant culture vessels | |||||||||
| Before application | apply dsRNA directly in the TC plant | Difference in Ct | ||||||
| Technical replicate 1 | Technical replicate 2 | Technical replicate 3 | Technical replicate 1 | Technical replicate 2 | Technical replicate 3 | Technical replicate 1 | Technical replicate 2 | Technical replicate 3 |
replicate 1 | 19.9 | 20.0 | 20.0 | 20.8 | 20.7 | 20.5 | 1.0 | 0.8 | 0.5 |
replicate 2 | 19.9 | 20.0 | 20.0 | 21.0 | 20.6 | 20.7 | 1.1 | 0.6 | 0.7 |
replicate 3 | 19.9 | 20.0 | 20.0 | 20.5 | 19.9 | 20.2 | 0.7 | -0.1 | 0.2 |
replicate 4 | 19.9 | 20.0 | 20.0 | 22.7 | 21.7 | 22.4 | 2.9 | 1.7 | 2.4 |
replicate 5 | 19.9 | 20.0 | 20.0 | 23.4 | 22.4 | 23.2 | 3.5 | 2.4 | 3.2 |
replicate 6 | 19.9 | 20.0 | 20.0 | 21.2 | 21.4 | 21.2 | 1.3 | 1.4 | 1.2 |
replicate 7 | 19.9 | 20.0 | 20.0 | 20.4 | 19.9 | 20.3 | 0.5 | -0.1 | 0.3 |
replicate 8 | 19.9 | 20.0 | 20.0 | 19.7 | 19.8 | 19.6 | -0.2 | -0.2 | -0.4 |
replicate 9 | 19.9 | 20.0 | 20.0 | 20.6 | 20.9 | 20.1 | 0.7 | 0.9 | 0.1 |
| Average = 20.0 | Standard deviation = 0.1 | Average = 21.0 | Standard deviation = 1.1 | Average = 1.0 | Standard deviation = 1.1 | Significance level (P) > 0.10 | ||
| Before application | apply the second PCR product directly in the TC plant | Difference in Ct | ||||||
| Technical replicate 1 | Technical replicate 2 | Technical replicate 3 | Technical replicate 1 | Technical replicate 2 | Technical replicate 3 | Technical replicate 1 | Technical replicate 2 | Technical replicate 3 |
replicate 1 | 19.2 | 19.6 | 19.6 | 19.3 | 20.0 | 19.7 | 0.1 | 0.4 | 0.1 |
replicate 2 | 19.2 | 19.6 | 19.6 | 19.4 | 19.0 | 19.7 | 0.2 | -0.6 | 0.1 |
replicate 3 | 19.2 | 19.6 | 19.6 | 19.3 | 19.7 | 20.6 | 0.1 | 0.2 | 1.0 |
replicate 4 | 19.2 | 19.6 | 19.6 | 19.2 | 19.9 | 19.7 | 0.0 | 0.3 | 0.1 |
replicate 5 | 19.2 | 19.6 | 19.6 | 19.6 | 19.0 | 20.0 | 0.5 | -0.6 | 0.4 |
replicate 6 | 19.2 | 19.6 | 19.6 | 19.9 | 19.8 | 20.8 | 0.7 | 0.2 | 1.2 |
replicate 7 | 19.2 | 19.6 | 19.6 | 19.6 | 19.3 | 19.1 | 0.4 | -0.3 | -0.5 |
replicate 8 | 19.2 | 19.6 | 19.6 | 19.8 | 19.6 | 20.5 | 0.6 | 0.0 | 0.9 |
replicate 9 | 19.2 | 19.6 | 19.6 | 19.9 | 19.8 | 19.5 | 0.7 | 0.2 | -0.2 |
| Average = 19.6 | Standard deviation = 0.2 | Average = 19.7 | Standard deviation = 0.4 | Average = 0.1 | Standard deviation = 0.4 | Significance level (P) > 0.10 | ||
replicate 1 | 20.5 | 20.1 | 20.2 | 20.1 | 20.8 | 20.8 | -0.4 | 0.7 | 0.7 |
replicate 2 | 20.5 | 20.1 | 20.2 | 21.1 | 20.9 | 20.7 | 0.7 | 0.8 | 0.5 |
replicate 3 | 20.5 | 20.1 | 20.2 | 19.9 | 20.4 | 20.1 | -0.5 | 0.2 | -0.1 |
replicate 4 | 20.5 | 20.1 | 20.2 | 19.9 | 20.7 | 20.4 | -0.6 | 0.6 | 0.2 |
replicate 5 | 20.5 | 20.1 | 20.2 | 21.0 | 20.7 | 20.6 | 0.5 | 0.6 | 0.4 |
replicate 6 | 20.5 | 20.1 | 20.2 | 20.6 | 20.3 | 20.2 | 0.2 | 0.2 | 0.1 |
replicate 7 | 20.5 | 20.1 | 20.2 | 20.3 | 20.2 | 19.9 | -0.1 | 0.0 | -0.3 |
replicate 8 | 20.5 | 20.1 | 20.2 | 20.5 | 20.3 | 20.5 | 0.0 | 0.2 | 0.3 |
replicate 9 | 20.5 | 20.1 | 20.2 | 21.3 | 22.3 | 22.0 | 0.9 | 2.2 | 1.8 |
| Average = 20.2 | Standard deviation = 0.2 | Average = 20.6 | Standard deviation = 0.6 | Average = 0.4 | Standard deviation = 0.6 | Significance level (P) 0.10 > P > 0.05 | ||
| Before application | no treatment | Difference in Ct | ||||||
| Technical replicate 1 | Technical replicate 2 | Technical replicate 3 | Technical replicate 1 | Technical replicate 2 | Technical replicate 3 | Technical replicate 1 | Technical replicate 2 | Technical replicate 3 |
replicate 1 | 19.6 | 19.8 | 20.0 | 19.4 | 19.2 | 20 | -0.2 | -0.6 | 0 |
replicate 2 | 19.6 | 19.8 | 20.0 | 20.0 | 20.4 | 20.5 | 0.4 | 0.5 | 0.5 |
replicate 3 | 19.6 | 19.8 | 20.0 | 19.7 | 20.6 | 20.3 | 0.1 | 0.7 | 0.3 |
replicate 4 | 19.6 | 19.8 | 20.0 | 19.9 | 20.3 | 20.6 | 0.3 | 0.5 | 0.7 |
replicate 5 | 19.6 | 19.8 | 20.0 | 20.1 | 19.9 | 20.3 | 0.5 | 0.1 | 0.4 |
replicate 6 | 19.6 | 19.8 | 20.0 | 19.2 | 20.1 | 20.1 | -0.4 | 0.2 | 0.1 |
replicate 7 | 19.6 | 19.8 | 20.0 | 20.3 | 20.6 | 20.8 | 0.7 | 0.8 | 0.8 |
replicate 8 | 19.6 | 19.8 | 20.0 | 20.5 | 20.5 | 20.7 | 0.9 | 0.6 | 0.8 |
replicate 9 | 19.6 | 19.8 | 20.0 | 20.1 | 20.0 | 20.2 | 0.5 | 0.2 | 0.2 |
| Average = 19.8 | Standard deviation= 0.2 | Average = 20.2 | Standard deviation = 0.4 | Average = 0.4 | Standard deviation = 0.4 | Significance level (P) > 0.10 | ||
Table S7: The average cycle threshold (Ct) values with standard deviation comparing short synthetic grapevine pinot gris virus (GPGV) targeted dsRNA treatment with other controls by either dipping tissue culture (TC) grapevine shoot tips for 2 or 24 hours prior to reintroduction into tissue culture or directly applying treatments to the leaves of plantlets in tissue culture.
Treatment | Average | Standard deviation | Treatment | Average | Standard deviation | Difference in Ct | p-value |
dip in dsRNA for 2 hrs | 23.9 | 0.9 | dip in the second PCR product for 2 hrs | 20.2 | 0.5 | -3.7 | P < 0.0001 |
dip in dsRNA for 2 hrs | 23.9 | 0.9 | dip in buffer control (water) for 2 hrs | 21.2 | 0.5 | -2.7 | P < 0.0001 |
dip in dsRNA for 2 hrs | 23.9 | 0.9 | no treatment | 21.8 | 0.8 | -2.1 | P < 0.0001 |
dip in dsRNA for 24 hrs | 24.9 | 1.2 | dip in the second PCR product for 22 hrs | 21.7 | 0.6 | -3.2 | P < 0.0001 |
dip in dsRNA for 24 hrs | 24.9 | 1.2 | dip in buffer control (water) for 24 hrs | 19.8 | 0.5 | -5.1 | P < 0.0001 |
dip in dsRNA for 24 hrs | 24.9 | 1.2 | no treatment | 20.9 | 0.5 | -4.0 | P < 0.0001 |
apply dsRNA directly in the TC plant | 21.0 | 1.1 | apply the second PCR product directly in the TC plant | 19.7 | 0.4 | -1.3 | P < 0.0001 |
apply dsRNA directly in the TC plant | 21.0 | 1.1 | apply buffer control (water) directly in the TC plant | 20.6 | 0.6 | -0.4 | P > 0.10 |
apply dsRNA directly in the TC plant | 21.0 | 1.1 | no treatment | 20.2 | 0.4 | -0.8 | 0.10 > P > 0.05 |
Table S8: The average cycle threshold (Ct) values with standard deviation comparing short synthetic grapevine pinot gris virus (GPGV) targeted dsRNA treatment with other controls by either dipping tissue culture (TC) grapevine shoot tips for 2 or 24 hours prior to reintroduction into tissue culture or directly applying treatments to the leaves of plantlets in tissue culture.
Treatment | Average | Standard deviation | Treatment | Average | Standard deviation | Difference in Ct | p-value |
dip in dsRNA for 2 hrs | 23.9 | 0.9 | dip in dsRNA for 24 hrs | 24.9 | 1.2 | 1.0 | 0.10 > P > 0.05 |
dip in dsRNA for 2 hrs | 23.9 | 0.9 | apply dsRNA directly in the TC plant | 21.0 | 1.1 | -2.9 | P < 0.0001 |
dip in dsRNA for 24 hrs | 24.9 | 1.2 | apply dsRNA directly in the TC plant | 21.0 | 1.1 | -3.9 | P < 0.0001 |
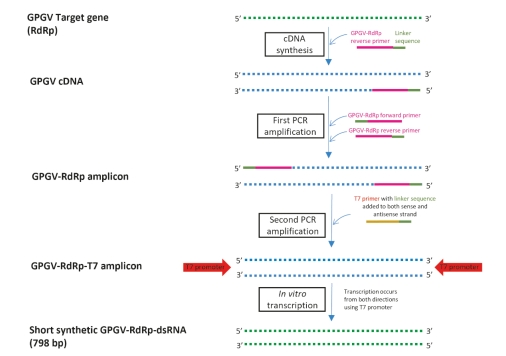
Figure S1: In vitro short synthetic grapevine Pinot gris virus (GPGV) double-stranded RNA (dsRNA) production using reverse transcription (RT) for cDNA synthesis and two-step PCR amplification of the GPGV RNA dependent RNA polymerase (RDRP) target sequence. A linker sequence is added to the reverse transcribed single-stranded copy DNA (cDNA) and to the 5’ ends of the double-stranded GPGV-RDRP amplicons in the first step of PCR amplification. The T7 promoter is added during the second step of PCR amplification. The derived GPGV-RdRp-T7 amplicon with the T7 prompter sequence is used for in vitro transcription to form the final short synthetic GPGV dsRNA.
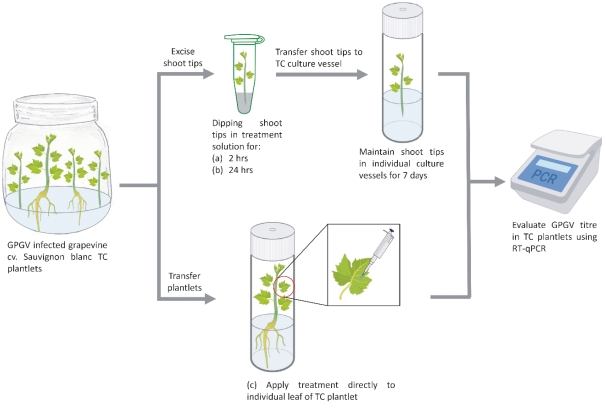
Figure S2: Exogenous application of short synthetic grapevine Pinot gris virus RNA dependent RNA polymerase double stranded RNA (GPGV- RdRp-dsRNA) or GPGV-RdRp-T7 amplicon or buffer control treatments to GPGV infected grapevine cv. Sauvignon Blanc tissue culture (TC) plantlet materials using three application methods: (a) dipping shoot tips excised from TC plants into GPGV-RdRp–dsRNA or control solutions for 2 hrs; (b) dipping shoot tips excised from TC plants into short synthetic GPGV dsRNA or control solutions for 24 hrs; and (c) exogenously applied GPGV-RdRp–dsRNA or control solutions to a single leaf of a TC plantlet in a culture vessel with media (Source: Biorender).
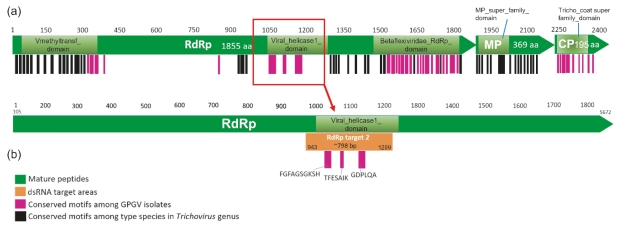
Figure S3: Schematic diagram indicating the conserved amino acid motif positions amongst a) Six exemplar species within the genus Trichovirus and their location on the amino acid sequences for the RNA-dependent RNA polymerase (RdRp; NCBI Reference Sequence: YP_004732978.2), movement protein (MP; NCBI Reference Sequence: YP_004732979.2) and coat protein (CP; NCBI Reference Sequence: YP_004732980.2) derived from the exemplar GPGV isolate (Refseq: NC_015782.2) and b) the enlarged view of the RdRp protein sequence illustrating the conserved amino acid motif positions amongst the 200 GPGV isolates and associated 798 bp target region to which the short synthetic GPGV dsRNA used to stimulate RNAi against GPGV was designed. The numbers indicate amino acid positions.
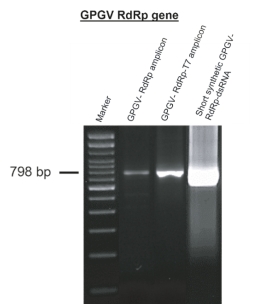
Figure S4. Visualization by agarose gel electrophoresis of the grapevine Pinot gris virus (GPGV) RNA-dependent RNA polymerase (RdRp) amplicons: GPGV-RdRp amplicon, GPGV-RdRp-T7 amplicon and the 798-base pair short synthetic GPGV-RdRp-double stranded RNA (GPGV-RdRp-dsRNA) molecule that was used to target the conserved 798 nucleotide region of the RdRp gene of GPGV to stimulate RNA interference and reduce GPGV titre. The polymerase chain reaction (PCR) amplicons and dsRNA molecules were produced after the two-step PCR method and in-vitro dsRNA transcription. Marker: 100 bp DNA ladder.